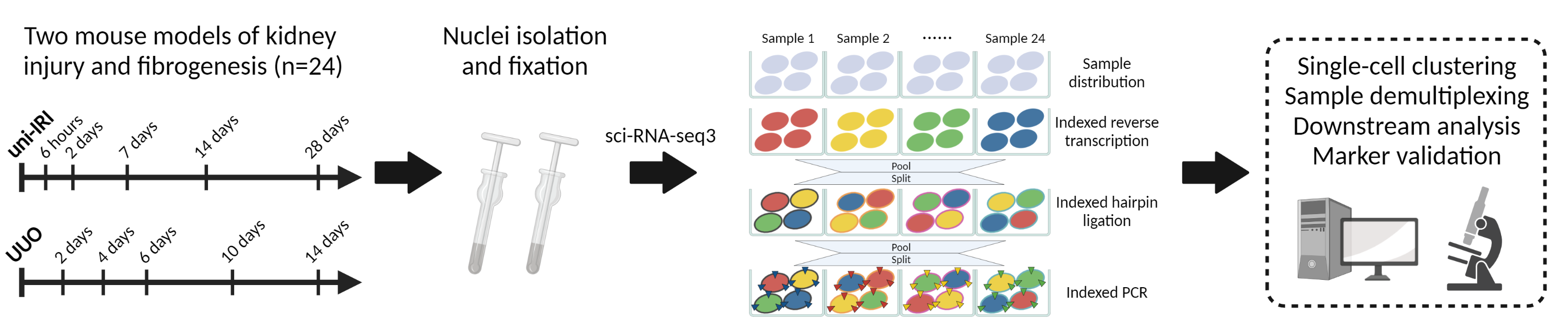
Comprehensive single-cell transcriptional profiling defines epithelial injury responses during kidney fibrogenesis
This repository documents the scripts to generate data for our manuscript studying mouse kidney fibrogenesis with sci-RNA-seq3.
The work is published on Cell Metabolism in 2022.
For citation (DOI: https://doi.org/10.1016/j.cmet.2022.09.026) (PMID: 36265491):
Li, H., Dixon, E. E., Wu, H., & Humphreys, B. D. (2022). Comprehensive single-cell transcriptional profiling defines shared and unique epithelial injury responses during kidney fibrosis. Cell metabolism, 34(12), 1977-1998.
Our detailed step-by-step protocol of sci-RNA-seq library generation can be found here at STAR Protocols.
Li, H., & Humphreys, B. D. (2022). Mouse kidney nuclear isolation and library preparation for single-cell combinatorial indexing RNA sequencing. STAR protocols, 3(4), 101904.
DOI: https://doi.org/10.1016/j.xpro.2022.101904 (PMID: 36595916)
The raw (.fastq) and pre-processed (count matrix) data and metadata have been deposited in NCBI’s Gene Expression Omnibus and are available through GEO Series accession number GSE190887.
A searchable database, including gene expression in all kidney cell types and PT cell types is available at our Kidney Interactive Transcriptomics (K.I.T.) website: http://humphreyslab.com/SingleCell/.
Pre-processing of raw fastq files was performed as previously described (Cao et al. Nature 2019; Cao et al. Science 2020): https://github.com/JunyueC/sci-RNA-seq3_pipeline
Quality control and single-cell clustering
Pseudobulk trajectory ordering (Fig. 1c)
Single-cell visualization (Fig. 1d)
Gene expression visualization (Fig. 1e)
Single-cell visualization (Fig. 2a)
Gene expression visualization (Fig. 2b)
Single-cell TF activity analysis (Fig. 2c)
Single-cell pathway activity analysis (Fig. 2d)
Single-cell trajectory ordering (Fig. 2f)
Fate prediction analysis with time-series scRNAseq data (Fig. 2g)
Quality control and single-cell subclustering
Single-cell visualization (Fig. 6a/b)
Gene expression visualization (Fig. 6c/d/f)
Quality control and single-cell subclustering
Single-cell visualization (Fig. 7a/b)
Gene module scoring (Fig. 7e)
Commands used for CellPhoneDB analysis
Statistical analysis (Fig. 7f/g)
RT barcode - Sample Lookup Table
Meta info of all cells used for clustering
**************
Find us on Twitter:
@HumphreysLab