The Superresolution Emitter Counter, as used to perform some analysis steps of the following publication:
Matamouros S., Gensch T., Cerff M., Sachs C.C., Abdoullahzadeh I., Hendriks J., Horst L., Tenhaef N., Noack, S., Graf M., Takors R., Nöh K. and Bott M.
Growth-rate dependency of ribosome abundance and translation elongation rate in Corynebacterium glutamicum differs from Escherichia coli
Submitted. Preprint available at bioRxiv: 10.1101/2021.04.01.438067
The analysis routines are tailored to the use case of the publication. It may be necessary to perform adaptations for other uses. Please cite our publication in case you use SurEmCo.
SurEmCo was tested under Windows 7 and 10, as well as Ubuntu Linux 18.04 and 20.04 (and should work under different distributions as well).
SurEmCo is written in Python 3 and relies on libraries as configured in setup.py. The library versions tested are defined in the environment.yml Anaconda environment.
We recommend using the Anaconda Python distribution. The installation will take roughly 5-15 minutes, depending on internet connection speed for Anaconda package download.
The conda-forge channel is needed, the suremco packages resides in the modsim channel:
conda config --add channels conda-forge # add conda-forge channel if not already present
conda install -yc modsim suremco
Alternatively, an environment can be created as defined in the environment.yml as part of this repository to install a set of known-working dependency versions:
conda env create -f environment.yml
conda activate suremco
To install SurEmCo from source, the tracker needs to be built, which was only tested with Ubuntu Linux (among others, the g++ g++-mingw-w64-x86-64 packages are needed), where it is cross-compiled for Windows as well. Run python setup.py install.
SurEmCo can then be started via
python -m suremco
# or with filenames
python -m suremco dia_image.png snsmil_output.txt
For testing SurEmCo, we provide a dataset taken from the study, available under the demodataset branch of this repository. You can download the whole demo dataset zipped here.
You can then either run SurEmCo without parameters (python -m suremco) and select all files (.txt as well as the .png), or directly start SurEmCo with the files as argument (e.g. python -m suremco demodataset/*).
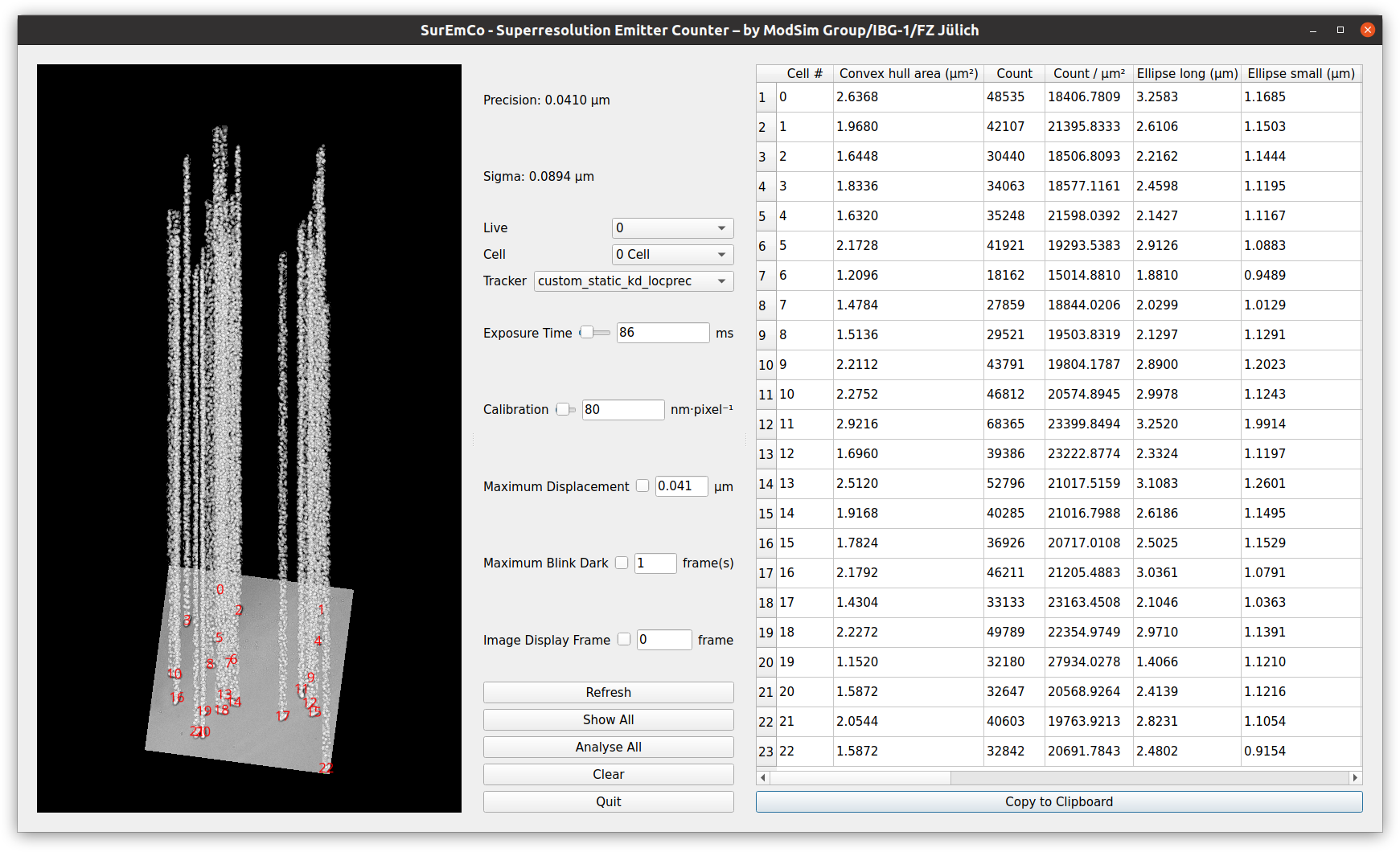
The software will load all emitter positions from the text file, and then show a GUI, this process should take less than a minute to complete on an average personal computer.
Certain parameters affect the results. Appropriate parameters for demodataset are: Exposure time 86 ms, Calibration 80 nm/pixel, Maximum displacement 0.041 µm, Maximum Blink dark 1 frame. Image Display Frame is for visual inspection in the 3D view only, and does not affect values.
After pressing the button Analyse All, all detected cells (as shown in the 2D/3D view) are analysed. For the demodataset, this process should again take less than a minute. The values should then match those in the screenshot, additionally showing a 3D view on the left denoting emitters (time as z dimension) of cells, with tracking results visualized by lines connecting emitters.
It is highly recommended to use the latest version of SurEmCo from Anaconda or this repository. To reproduce the exact results in the publication, use the version tagged as usedForAnalyses.
BSD