-
Notifications
You must be signed in to change notification settings - Fork 5
Prostate Example
Michel Lang edited this page Nov 11, 2013
·
4 revisions
- Prostate dataset from ElemStatLearn package
- Column "lpsa" is continous response
- Column "train" is originally used split into train/test
- Test if the split into train and test set is "lucky": compare with models based on fit on a random split
Source as R-markdown: Prostate-Example.Rmd.
library(BatchExperiments)
library(ElemStatLearn)
library(glmnet)
library(plyr)
# Load data and get a quick overview
data("prostate", package = "ElemStatLearn")
str(prostate)## 'data.frame': 97 obs. of 10 variables:
## $ lcavol : num -0.58 -0.994 -0.511 -1.204 0.751 ...
## $ lweight: num 2.77 3.32 2.69 3.28 3.43 ...
## $ age : int 50 58 74 58 62 50 64 58 47 63 ...
## $ lbph : num -1.39 -1.39 -1.39 -1.39 -1.39 ...
## $ svi : int 0 0 0 0 0 0 0 0 0 0 ...
## $ lcp : num -1.39 -1.39 -1.39 -1.39 -1.39 ...
## $ gleason: int 6 6 7 6 6 6 6 6 6 6 ...
## $ pgg45 : int 0 0 20 0 0 0 0 0 0 0 ...
## $ lpsa : num -0.431 -0.163 -0.163 -0.163 0.372 ...
## $ train : logi TRUE TRUE TRUE TRUE TRUE TRUE ...
pairs(prostate, col = c("red", "darkgreen")[prostate$train + 1])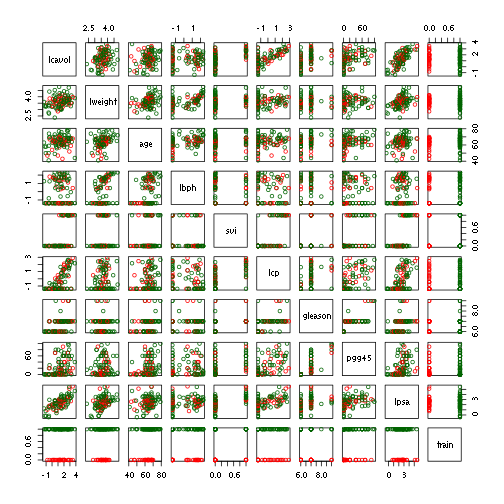
# Create registry
file.dir <- tempfile("prostate_")
reg <- makeExperimentRegistry("prostate_example", file.dir = file.dir, packages = "glmnet",
seed = 1)## Creating dir: /tmp/RtmpbOS7J6/prostate_23583d1eadeb
## Saving registry: /tmp/RtmpbOS7J6/prostate_23583d1eadeb/registry.RData
# we explicitly use the multicore backend here, regardless of what you have
# in your config
setConfig(conf = list(cluster.functions = makeClusterFunctionsMulticore(4)))
# Define subsample function
ss <- function(static, orig = FALSE) {
nr <- nrow(static)
train.ind <- if (orig)
static$train else sample(static$train)
list(train = subset(static, train.ind, select = -train), test = subset(static,
!train.ind, select = -train))
}
# Function to fit a simple linear model
linear <- function(dynamic) {
model <- lm(lpsa ~ ., data = dynamic$train)
y <- dynamic$test$lpsa
y.hat <- predict(model, newdata = dynamic$test)
1 - sum((y - y.hat)^2)/sum((y - mean(y))^2) # R^2
}
# Function to fit a regularized regression model, see ?glmnet
regularized <- function(dynamic, alpha) {
x <- as.matrix(subset(dynamic$train, select = -lpsa))
y <- dynamic$train[["lpsa"]]
model <- cv.glmnet(x, y, alpha = alpha)
x <- as.matrix(subset(dynamic$test, select = -lpsa))
y <- dynamic$test$lpsa
y.hat <- predict(model, newx = x)
1 - sum((y - y.hat)^2)/sum((y - mean(y))^2) # R^2
}
# Add problem and algorithms to the registry
addProblem(reg, "prostate", static = prostate, dynamic = ss, seed = 1)## Writing problem files: /tmp/RtmpbOS7J6/prostate_23583d1eadeb/problems/prostate_static.RData, /tmp/RtmpbOS7J6/prostate_23583d1eadeb/problems/prostate_dynamic.RData
addAlgorithm(reg, "linear", fun = linear)## Writing algorithm file: /tmp/RtmpbOS7J6/prostate_23583d1eadeb/algorithms/linear.RData
addAlgorithm(reg, "lasso", fun = regularized)## Writing algorithm file: /tmp/RtmpbOS7J6/prostate_23583d1eadeb/algorithms/lasso.RData
addAlgorithm(reg, "ridge", fun = regularized)## Writing algorithm file: /tmp/RtmpbOS7J6/prostate_23583d1eadeb/algorithms/ridge.RData
# Add experiments using the original split
pd <- makeDesign("prostate", design = data.frame(orig = TRUE))
ad <- list(makeDesign("linear"), makeDesign("lasso", design = data.frame(alpha = 1)),
makeDesign("ridge", design = data.frame(alpha = 0)))
addExperiments(reg, pd, ad)## Adding 3 experiments / 3 jobs to DB.
# Add experiments using random splits, 100 replications
pd <- makeDesign("prostate", design = data.frame(orig = FALSE))
addExperiments(reg, pd, ad, repl = 100)## Adding 3 experiments / 300 jobs to DB.
# Quick overview: defined experiments
summarizeExperiments(reg)## prob algo .count
## 1 prostate lasso 101
## 2 prostate linear 101
## 3 prostate ridge 101
# Chunk jobs together - they are pretty fast
chunked <- chunk(getJobIds(reg), n.chunks = 10, shuffle = TRUE)
submitJobs(reg, chunked)## Saving conf: /tmp/RtmpbOS7J6/prostate_23583d1eadeb/conf.RData
## Submitting 10 chunks / 303 jobs.
## Cluster functions: Multicore.
## Auto-mailer settings: start=none, done=none, error=none.
## Writing 10 R scripts...
## Sending 10 submit messages...
## Might take some time, do not interrupt this!
## 2 temporary submit errors logged to file '/tmp/RtmpbOS7J6/prostate_23583d1eadeb/submit.log'.
## First message: Multicore busy: J
# Check computational status
showStatus(reg)## Status for 303 jobs at 2013-11-11 12:55:02
## Submitted: 303 (100.00%)
## Started: 242 ( 79.87%)
## Running: 0 ( 0.00%)
## Done: 242 ( 79.87%)
## Errors: 0 ( 0.00%)
## Expired: 0 ( 0.00%)
## Time: min=0.00s avg=0.12s max=1.00s
# Wait for the jobs first
waitForJobs(reg)## [1] TRUE
# Get grouped mean values
df <- reduceResultsExperiments(reg, fun = function(job, res) list(R2 = res))## Reducing 303 results...
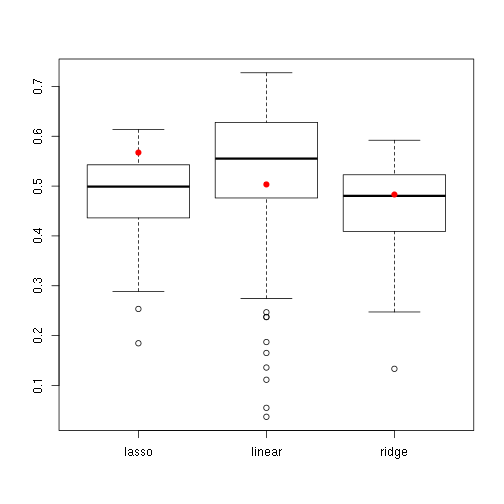
ddply(df, c("orig", "algo"), summarise, n = length(R2), mean = mean(R2))## orig algo n mean
## 1 FALSE lasso 100 0.4831
## 2 FALSE linear 100 0.5235
## 3 FALSE ridge 100 0.4634
## 4 TRUE lasso 1 0.5674
## 5 TRUE linear 1 0.5034
## 6 TRUE ridge 1 0.4831
# Extract R2 of original split
ids <- findExperiments(reg, prob.pars = (orig == TRUE))
df <- reduceResultsExperiments(reg, ids, fun = function(job, res) list(R2 = res))## Reducing 3 results...
orig <- unlist(split(df$R2, df$algo))
# Extract R2 of subsampled splits
ids <- findExperiments(reg, prob.pars = (orig == FALSE))
df <- reduceResultsExperiments(reg, ids, fun = function(job, res) list(R2 = res))## Reducing 300 results...
subsampled <- as.data.frame(split(df$R2, df$algo))
# boxplot of the results
boxplot(subsampled)
points(1:3, orig[names(subsampled)], col = "red", pch = 19)